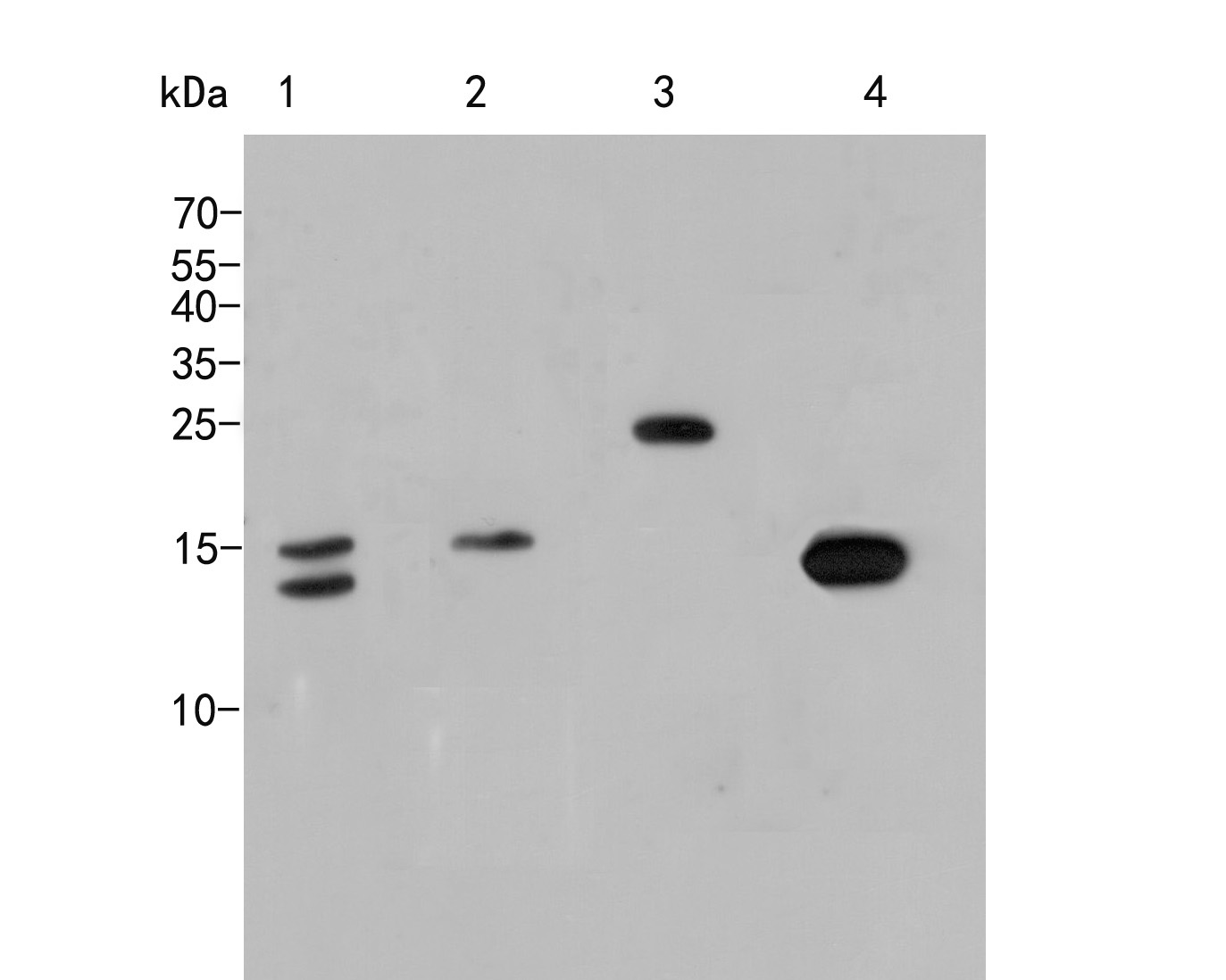
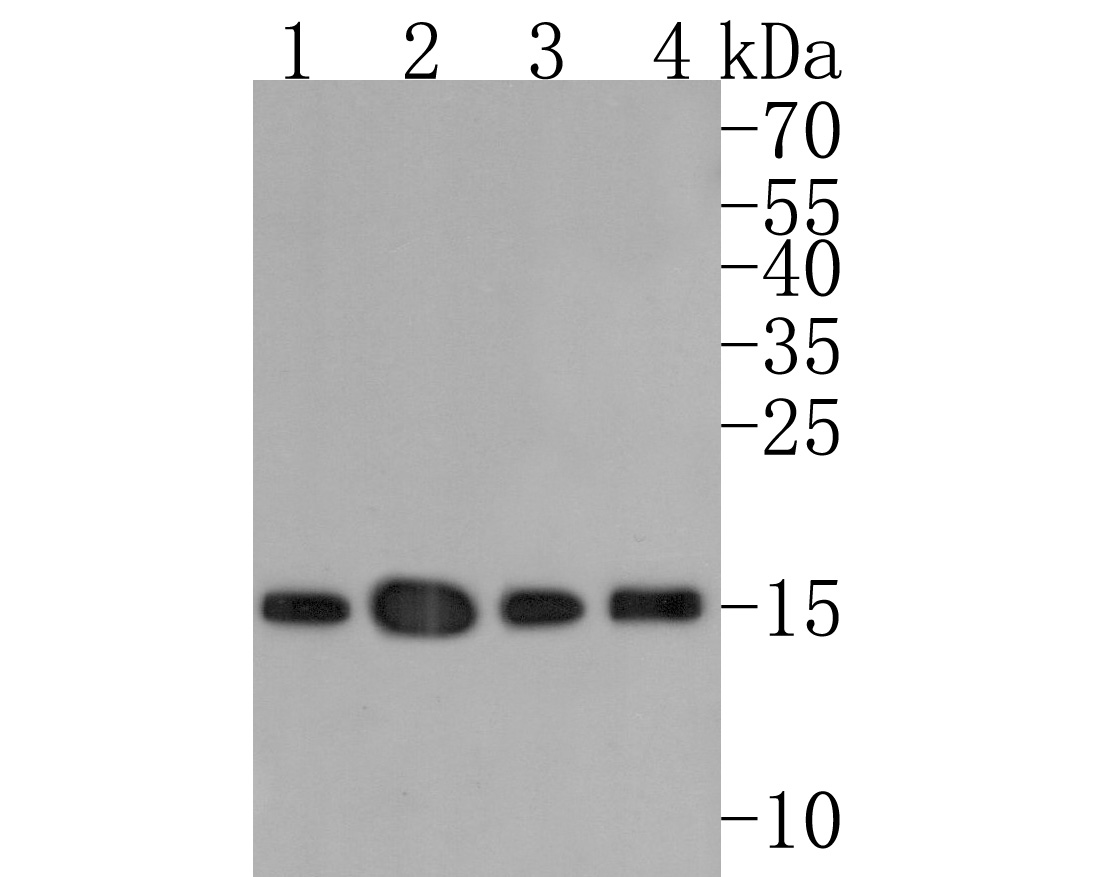
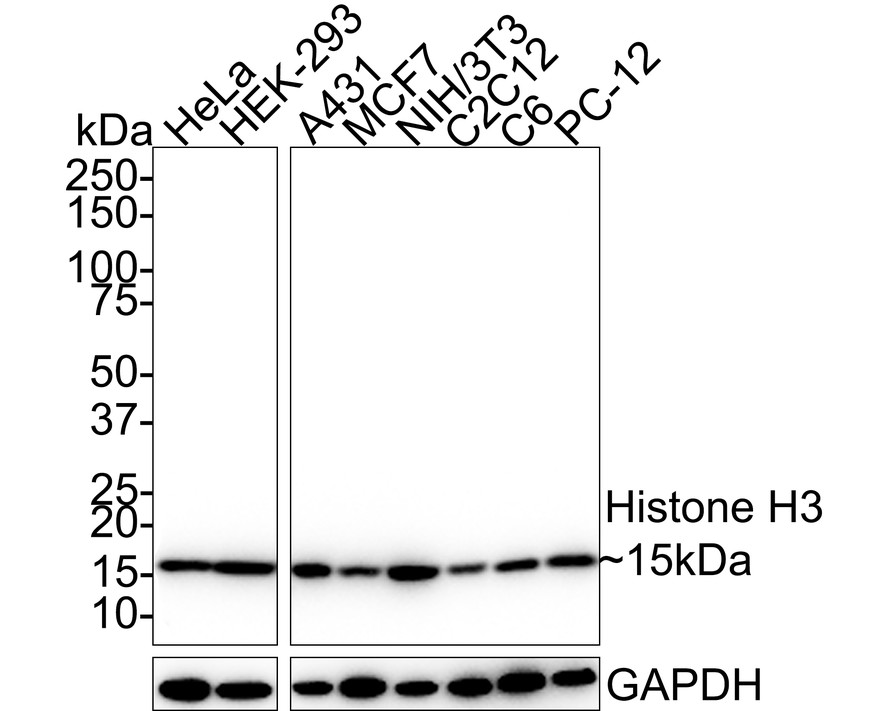
概述
产品名称
Histone H3 Rabbit Polyclonal Antibody
抗体类型
Rabbit Polyclonal Antibody
免疫原
Synthetic peptide within human Histone H3 aa 1-50.
种属反应性
Human, Mouse, Rat
验证应用
WB
分子量
Predicted band size: 15 kDa
阳性对照
HepG2, NIH/3T3, Rat liver
偶联
unconjugated
RRID
产品特性
形态
Liquid
浓度
1ug/ul
存放说明
Store at +4℃ after thawing. Aliquot store at -20℃ or -80℃. Avoid repeated freeze / thaw cycles.
存储缓冲液
1*PBS (pH7.4), 0.2% BSA, 40% Glycerol. Preservative: 0.05% Sodium Azide.
亚型
IgG
纯化方式
Immunogen affinity purified.
应用稀释度
-
WB
-
1:1,000~1:2,000
发表文章中的应用
发表文章中的种属
| Pig | See 2 publications below |
| human | See 2 publications below |
| Embryonic Stem Cells | See 1 publications below |
靶点
功能
The nucleosome, made up of DNA wound around eight core histone proteins (two each of H2A, H2B, H3, and H4), is the primary building block of chromatin. Histones thereby play a central role in transcription regulation, DNA repair, DNA replication and chromosomal stability. DNA accessibility is regulated via a complex set of post-translational modifications of histones, also called histone code, and nucleosome remodeling. Histone H3 is primarily acetylated at Lys9, 14, 18, 23, 27, and 56. Acetylation of H3 at Lys9 appears to have a dominant role in histone deposition and chromatin assembly in some organisms. This Antibody detects endogenous levels of total histone H3 protein, and does not cross-react with other histones.
背景文献
1. Flanagan J.F., Mi L.-Z., Chruszcz M., Cymborowski M., Clines K.L., Kim Y., Minor W., Rastinejad F., Khorasanizadeh S."Double chromodomains cooperate to recognize the methylated histone H3 tail."Nature 438:1181-1185(2005)
2. "Arginine methylation of the histone H3 tail impedes effector binding."Iberg A.N., Espejo A., Cheng D., Kim D., Michaud-Levesque J., Richard S., Bedford M.T.J. Biol. Chem. 283:3006-3010(2008)
序列相似性
Belongs to the histone H3 family.
翻译后修饰
Acetylation is generally linked to gene activation. Acetylation on Lys-10 (H3K9ac) impairs methylation at Arg-9 (H3R8me2s). Acetylation on Lys-19 (H3K18ac) and Lys-24 (H3K24ac) favors methylation at Arg-18 (H3R17me). Acetylation at Lys-123 (H3K122ac) by EP300/p300 plays a central role in chromatin structure: localizes at the surface of the histone octamer and stimulates transcription, possibly by promoting nucleosome instability.; Citrullination at Arg-9 (H3R8ci) and/or Arg-18 (H3R17ci) by PADI4 impairs methylation and represses transcription.; Asymmetric dimethylation at Arg-18 (H3R17me2a) by CARM1 is linked to gene activation. Symmetric dimethylation at Arg-9 (H3R8me2s) by PRMT5 is linked to gene repression. Asymmetric dimethylation at Arg-3 (H3R2me2a) by PRMT6 is linked to gene repression and is mutually exclusive with H3 Lys-5 methylation (H3K4me2 and H3K4me3). H3R2me2a is present at the 3' of genes regardless of their transcription state and is enriched on inactive promoters, while it is absent on active promoters.; Methylation at Lys-5 (H3K4me), Lys-37 (H3K36me) and Lys-80 (H3K79me) are linked to gene activation. Methylation at Lys-5 (H3K4me) facilitates subsequent acetylation of H3 and H4. Methylation at Lys-80 (H3K79me) is associated with DNA double-strand break (DSB) responses and is a specific target for TP53BP1. Methylation at Lys-10 (H3K9me) and Lys-28 (H3K27me) are linked to gene repression. Methylation at Lys-10 (H3K9me) is a specific target for HP1 proteins (CBX1, CBX3 and CBX5) and prevents subsequent phosphorylation at Ser-11 (H3S10ph) and acetylation of H3 and H4. Methylation at Lys-5 (H3K4me) and Lys-80 (H3K79me) require preliminary monoubiquitination of H2B at 'Lys-120'. Methylation at Lys-10 (H3K9me) and Lys-28 (H3K27me) are enriched in inactive X chromosome chromatin. Monomethylation at Lys-57 (H3K56me1) by EHMT2/G9A in G1 phase promotes interaction with PCNA and is required for DNA replication.; Phosphorylated at Thr-4 (H3T3ph) by HASPIN during prophase and dephosphorylated during anaphase. Phosphorylation at Ser-11 (H3S10ph) by AURKB is crucial for chromosome condensation and cell-cycle progression during mitosis and meiosis. In addition phosphorylation at Ser-11 (H3S10ph) by RPS6KA4 and RPS6KA5 is important during interphase because it enables the transcription of genes following external stimulation, like mitogens, stress, growth factors or UV irradiation and result in the activation of genes, such as c-fos and c-jun. Phosphorylation at Ser-11 (H3S10ph), which is linked to gene activation, prevents methylation at Lys-10 (H3K9me) but facilitates acetylation of H3 and H4. Phosphorylation at Ser-11 (H3S10ph) by AURKB mediates the dissociation of HP1 proteins (CBX1, CBX3 and CBX5) from heterochromatin. Phosphorylation at Ser-11 (H3S10ph) is also an essential regulatory mechanism for neoplastic cell transformation. Phosphorylated at Ser-29 (H3S28ph) by MAP3K20 isoform 1, RPS6KA5 or AURKB during mitosis or upon ultraviolet B irradiation. Phosphorylation at Thr-7 (H3T6ph) by PRKCB is a specific tag for epigenetic transcriptional activation that prevents demethylation of Lys-5 (H3K4me) by LSD1/KDM1A. At centromeres, specifically phosphorylated at Thr-12 (H3T11ph) from prophase to early anaphase, by DAPK3 and PKN1. Phosphorylation at Thr-12 (H3T11ph) by PKN1 is a specific tag for epigenetic transcriptional activation that promotes demethylation of Lys-10 (H3K9me) by KDM4C/JMJD2C. Phosphorylation at Thr-12 (H3T11ph) by chromatin-associated CHEK1 regulates the transcription of cell cycle regulatory genes by modulating acetylation of Lys-10 (H3K9ac). Phosphorylation at Tyr-42 (H3Y41ph) by JAK2 promotes exclusion of CBX5 (HP1 alpha) from chromatin.; Monoubiquitinated by RAG1 in lymphoid cells, monoubiquitination is required for V(D)J recombination (By similarity). Ubiquitinated by the CUL4-DDB-RBX1 complex in response to ultraviolet irradiation. This may weaken the interaction between histones and DNA and facilitate DNA accessibility to repair proteins.; Lysine deamination at Lys-5 (H3K4all) to form allysine is mediated by LOXL2. Allysine formation by LOXL2 only takes place on H3K4me3 and results in gene repression.; Crotonylation (Kcr) is specifically present in male germ cells and marks testis-specific genes in post-meiotic cells, including X-linked genes that escape sex chromosome inactivation in haploid cells. Crotonylation marks active promoters and enhancers and confers resistance to transcriptional repressors. It is also associated with post-meiotically activated genes on autosomes.; Butyrylation of histones marks active promoters and competes with histone acetylation. It is present during late spermatogenesis.; Succinylation at Lys-80 (H3K79succ) by KAT2A takes place with a maximum frequency around the transcription start sites of genes. It gives a specific tag for epigenetic transcription activation. Desuccinylation at Lys-123 (H3K122succ) by SIRT7 in response to DNA damage promotes chromatin condensation and double-strand breaks (DSBs) repair.; Serine ADP-ribosylation constitutes the primary form of ADP-ribosylation of proteins in response to DNA damage. Serine ADP-ribosylation at Ser-11 (H3S10ADPr) is mutually exclusive with phosphorylation at Ser-11 (H3S10ph) and impairs acetylation at Lys-10 (H3K9ac).
亚细胞定位
Cell nucleus
别名
H3 histone family, member A antibody
H3/A antibody
H31_HUMAN antibody
H3FA antibody
Hist1h3a antibody
HIST1H3B antibody
HIST1H3C antibody
HIST1H3D antibody
HIST1H3E antibody
HIST1H3F antibody
展开H3 histone family, member A antibody
H3/A antibody
H31_HUMAN antibody
H3FA antibody
Hist1h3a antibody
HIST1H3B antibody
HIST1H3C antibody
HIST1H3D antibody
HIST1H3E antibody
HIST1H3F antibody
HIST1H3G antibody
HIST1H3H antibody
HIST1H3I antibody
HIST1H3J antibody
histone 1, H3a antibody
Histone cluster 1, H3a antibody
Histone H3.1 antibody
Histone H3/a antibody
Histone H3/b antibody
Histone H3/c antibody
Histone H3/d antibody
Histone H3/f antibody
Histone H3/h antibody
Histone H3/i antibody
Histone H3/j antibody
Histone H3/k antibody
Histone H3/l antibody
折叠图片
-
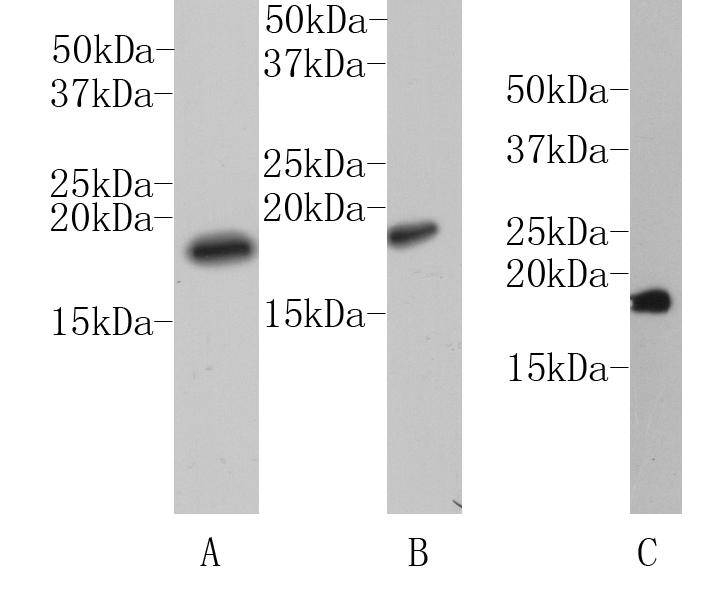
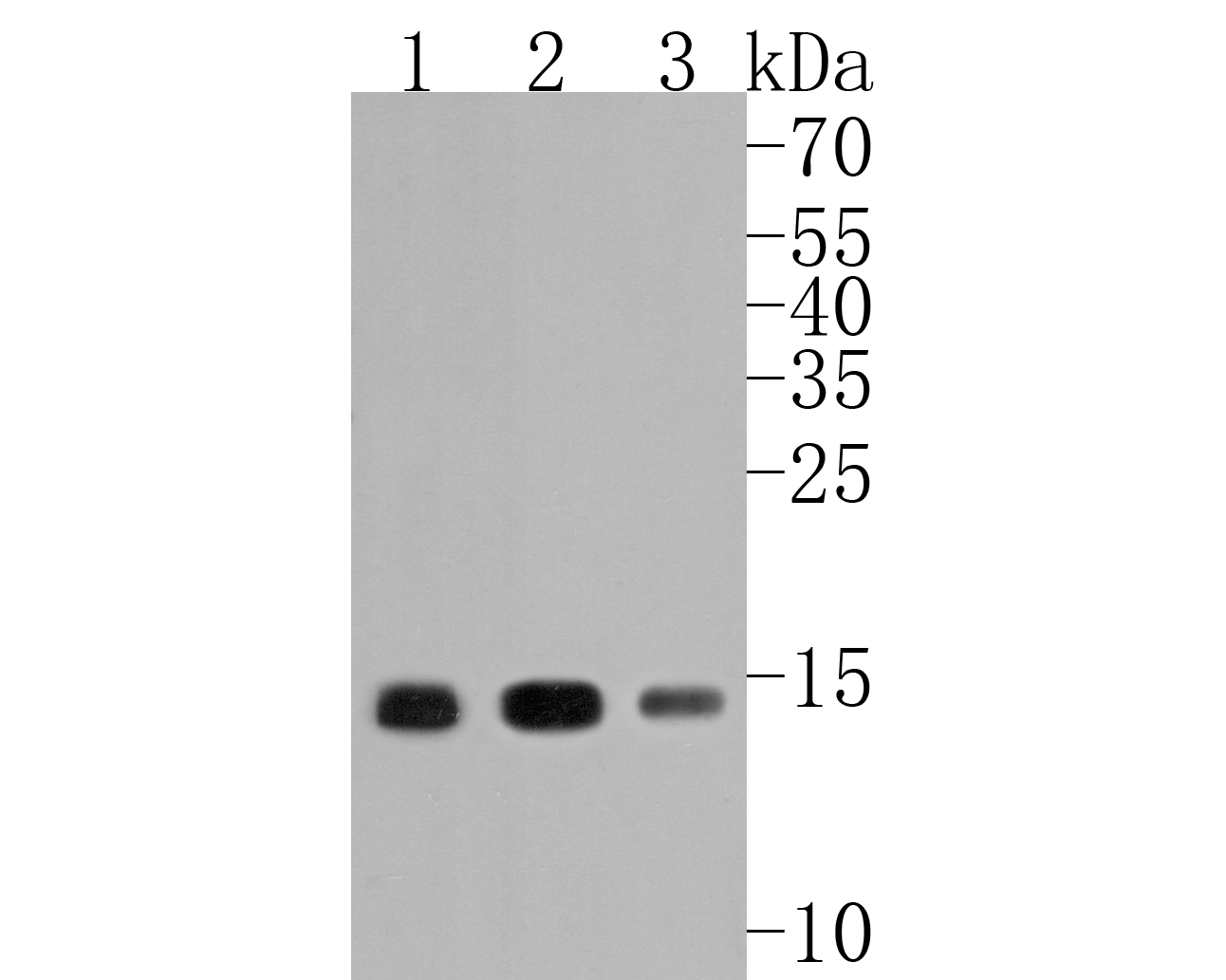
Western blot analysis on A:HepG2 B:NIH/3T3 C:Rat liver cell lysates using anti-Histone H3 polyclonal antibody.
Please note: All products are "FOR RESEARCH USE ONLY AND ARE NOT INTENDED FOR DIAGNOSTIC OR THERAPEUTIC USE"
引文
-
Contribution of DEAD-Box RNA Helicase 21 to the Nucleolar Localization of Porcine Circovirus Type 4 Capsid Protein
Author: Zhou, J., Wang, Y., Qiu, Y., Wang, Y., Yang, X., Liu, C., Shi, Y., Feng, X., Hou, L., & Liu, J.
PMID: 35283818
应用: WB
反应种属: Pig
发表时间: 2022 Feb
-
Citation
-
Porcine Circovirus Type 2 Hijacks Host IPO5 to Sustain the Intracytoplasmic Stability of Its Capsid Protein
Author: Lin, C., Hu, J., Dai, Y., Zhang, H., Xu, K., Dong, W., Yan, Y., Peng, X., Zhou, J., & Gu, J.
PMID: 36409110
应用:
反应种属:
发表时间: 2022 Dec
-
Citation
-
Nucleolar protein NPM1 is essential for circovirus replication by binding to viral capsid
Author: Jinyan Gu; Jiyong Zhou
PMID: 33073687
应用: WB,CoIP
反应种属: human
发表时间: 2020 Dec
-
Citation
-
Wnt Signaling Modulates Routes of Retinoic Acid-Induced Differentiation of Embryonic Stem Cells.
Author: Ming Zhang,Liangbiao Chen
PMID: 31337269
应用: WB,IS
反应种属: Embryonic Stem Cells
发表时间: 2019 Oct
-
Citation
-
Caspase-Dependent Apoptosis Induction via Viral Protein ORF4 of Porcine Circovirus 2 Binding to Mitochondrial Adenine Nucleotide Translocase 3
Author: Jiyong Zhou
PMID: 29491154
应用: WB
反应种属: Porcine Circovirus
发表时间: 2018 Apr
-
Citation
-
Cellular proteomic analysis of porcine circovirus type 2 and classical swine fever virus coinfection in porcine kidney‐15 cells using isobaric tags for relative and absolute quantitation-coupled LC-MS/MS
Author: Zhou, N., Fan, C., Liu, S., Zhou, J., Jin, Y., Zheng, X., Wang, Q., Liu, J., Yang, H., Gu, J., & Zhou, J.
PMID: 28247913
应用: WB
反应种属: Pig
发表时间: 2017 May
-
Citation
-
Circovirus Transport Proceeds via Direct Interaction of the Cytoplasmic Dynein IC1 Subunit with theViral Capsid Protein
Author: Jiyong Zhou
PMID: 25540360
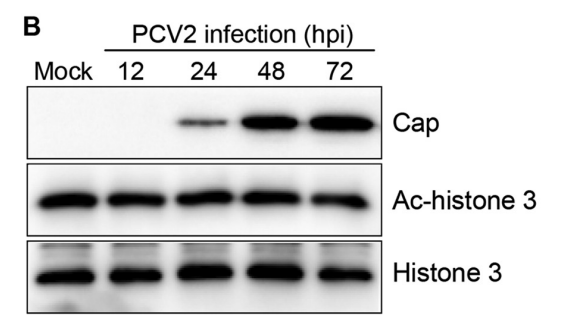
应用: WB,IP
反应种属: Circovirus
发表时间: 2015 Mar
-
Citation
-
Impaired Phosphorylation and Ubiquitination by p70 S6 Kinase (p70S6K) and Smad Ubiquitination Regulatory Factor1 (Smurf1) Promote Tribbles Homolog 2 (TRIB2) Stability and Carcinogenic Property in Liver Cancer
Author: Fenyong Sun
PMID: 24089522
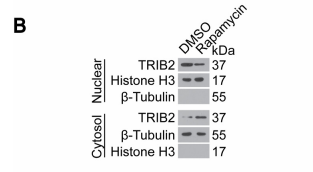
应用: WB
反应种属: human
发表时间: 2013 Nov
-
Citation
Alternative Products
Histone H3 Recombinant Rabbit Monoclonal Antibody [JJ092-08]
Application: WB,IF-Tissue,IHC-P,ChIP
Reactivity: Human,Mouse,Rat
Conjugate: unconjugated
Histone H3 Recombinant Rabbit Monoclonal Antibody [JJ090-07]
Application: WB,IF-Cell,IF-Tissue,IHC-P,ChIP,IP
Reactivity: Human,Mouse,Rat
Conjugate: unconjugated
同靶点&同通路的产品
Phospho-Histone H3 (S28) Recombinant Rabbit Monoclonal Antibody [JE56-06]
Application: WB,IF-Cell,IHC-P,FC,ChIP
Reactivity: Human,Rat,Mouse
Conjugate: unconjugated
Histone H3 (mono methyl K36) Recombinant Rabbit Monoclonal Antibody [SR04-20]
Application: WB,IF-Cell,IF-Tissue,ChIP
Reactivity: Human,Mouse
Conjugate: unconjugated
Histone H3 (mono methyl R2) Recombinant Rabbit Monoclonal Antibody [ST0427]
Application: WB,IF-Cell,IF-Tissue,IHC-P,ChIP
Reactivity: Human,Mouse,Rat
Conjugate: unconjugated
Histone H3 (acetyl K56) Recombinant Rabbit Monoclonal Antibody [SU30-10]
Application: WB,IF-Cell,IF-Tissue,IHC-P,ChIP,CUT&Tag-seq
Reactivity: Human,Mouse,Rat
Conjugate: unconjugated
Histone H3 (tri methyl K9) Mouse Monoclonal Antibody [1-6]
Application: WB,IF-Cell,IHC-P,IF-Tissue
Reactivity: Human,Mouse,Rat
Conjugate: unconjugated
Histone H3 Mouse Monoclonal Antibody [A11-D7]
Application: WB,IF-Cell,IHC-P,IF-Tissue
Reactivity: Human,Mouse,Rat,Zebrafish
Conjugate: unconjugated
Histone H3 (tri methyl K27) Recombinant Rabbit Monoclonal Antibody [PSH05-15]
Application: WB,IF-Cell,IHC-P,IF-Tissue,ChIP
Reactivity: Human,Mouse,Rat
Conjugate: unconjugated
Histone H3 (di methyl K4) Rabbit Polyclonal Antibody
Application: WB,IF-Cell,IHC-P,FC,Dot Blot
Reactivity: Human,Mouse,Rat
Conjugate: unconjugated
Histone H3 (acetyl K23) Recombinant Rabbit Monoclonal Antibody [JE46-39]
Application: WB,IF-Cell
Reactivity: Human,Mouse
Conjugate: unconjugated
Histone H3 Mouse Monoclonal Antibody [3-C4]
Application: WB,IHC-P,mIHC,IF-Tissue,ChIP
Reactivity: Human,Mouse,Rat
Conjugate: unconjugated
Histone H3 (mono methyl K18) Recombinant Rabbit Monoclonal Antibody [SA42-07]
Application: WB,IF-Cell,IF-Tissue,IHC-P
Reactivity: Human,Mouse,Rat
Conjugate: unconjugated
Phospho-Histone H3 (S10) Recombinant Rabbit Monoclonal Antibody [SA31-01]
Application: WB,IF-Cell,IF-Tissue,IHC-P,IP,FC,ChIP
Reactivity: Human,Mouse,Rat
Conjugate: unconjugated
Histone H3 (di methyl K9) Recombinant Rabbit Monoclonal Antibody [SN07-30]
Application: WB,IF-Cell,IF-Tissue,IHC-P
Reactivity: Human,Mouse,Rat
Conjugate: unconjugated
Histone H3 Recombinant Rabbit Monoclonal Antibody [JJ092-08]
Application: WB,IF-Tissue,IHC-P,ChIP
Reactivity: Human,Mouse,Rat
Conjugate: unconjugated
Histone H3 (acetyl K4) Recombinant Rabbit Monoclonal Antibody [PSH03-71]
Application: WB,IF-Cell,ChIP,Dot Blot
Reactivity: Human,Mouse,Rat
Conjugate: unconjugated
Histone H3 (acetyl K14) Recombinant Rabbit Monoclonal Antibody [JU43-26]
Application: WB,IF-Cell,IF-Tissue,IHC-P,IP,SNAP-ChIP,CUT&Tag-seq
Reactivity: Human,Mouse,Rat
Conjugate: unconjugated
Histone H3 (tri methyl K9) Mouse Monoclonal Antibody [2G1]
Application: WB,IF
Reactivity: Human
Conjugate: unconjugated
Histone H3 (acetyl K27) Rabbit Polyclonal Antibody
Application: WB,IHC-P,ChIP
Reactivity: Human,Mouse
Conjugate: unconjugated
Histone H3 (acetyl K18) Rabbit Polyclonal Antibody
Application: WB,IHC-P,IF-Cell
Reactivity: Human,Mouse
Conjugate: unconjugated
Histone H3 Rabbit Polyclonal Antibody
Application: WB
Reactivity: Zebrafish,Human,Mouse
Conjugate: unconjugated
Phospho-Histone H3 (T3) Recombinant Rabbit Monoclonal Antibody [JE42-48]
Application: WB,IHC-P
Reactivity: Human,Mouse,Rat
Conjugate: unconjugated
Histone H3 Mouse Monoclonal Antibody [6-A7]
Application: WB,IHC-P,IF-Tissue,ChIP
Reactivity: Human,Mouse,Rat
Conjugate: unconjugated
Histone H3 (mono methyl K4) Rabbit Polyclonal Antibody
Application: WB,IF-Cell,IHC-P,FC,Dot Blot
Reactivity: Human,Mouse,Rat
Conjugate: unconjugated
Histone H3 (acetyl K27) Mouse Monoclonal Antibody [A6D7]
Application: WB,IF-Cell,IHC-P,FC,ChIP
Reactivity: Human,Mouse,Rat
Conjugate: unconjugated
Histone H3 (acetyl K27) Mouse Monoclonal Antibody [A6D6]
Application: WB,IF-Cell,IHC-P,FC,ChIP
Reactivity: Human,Mouse,Rat
Conjugate: unconjugated
Histone H3 (acetyl K18) Mouse Monoclonal Antibody [A7B5]
Application: WB,IHC-P,ChIP
Reactivity: Human,Rat
Conjugate: unconjugated
Histone H3 Recombinant Mouse Monoclonal Antibody [A11-D7-R]
Application: WB,IF-Cell,IHC-P
Reactivity: Human,Mouse,Rat
Conjugate: unconjugated
Histone H3 Recombinant Mouse Monoclonal Antibody [6-A7-R] - Loading control
Application: WB,IHC-P,IF-Tissue,ChIP
Reactivity: Human,Mouse,Rat
Conjugate: unconjugated
Histone H3 Recombinant Rabbit Monoclonal Antibody [JJ090-07]
Application: WB,IF-Cell,IF-Tissue,IHC-P,ChIP,IP
Reactivity: Human,Mouse,Rat
Conjugate: unconjugated
Histone H3 (acetyl K9) Recombinant Rabbit Monoclonal Antibody [PSH04-47] - ChIP Grade
Application: WB,IF-Cell,IHC-P,IF-Tissue,FC,ChIP,Dot Blot,IP
Reactivity: Human,Mouse,Rat
Conjugate: unconjugated
Histone H3 (mono methyl K14) Recombinant Rabbit Monoclonal Antibody [JE43-29]
Application: WB,IF-Cell,ChIP,Dot Blot
Reactivity: Human,Mouse,Rat
Conjugate: unconjugated
Histone H3 (mono+di+tri methyl K79) Recombinant Rabbit Monoclonal Antibody [SR42-06]
Application: WB,IHC-P,ChIP
Reactivity: Human,Mouse,Rat
Conjugate: unconjugated
Histone H3 (acetyl K18) Mouse Monoclonal Antibody [A7B6]
Application: WB,IF-Cell
Reactivity: Human
Conjugate: unconjugated
Histone H3 (di methyl K4) Recombinant Rabbit Monoclonal Antibody [JE00-98]
Application: WB,IF-Cell,ChIP,Dot Blot,IHC-P
Reactivity: Human,Mouse,Rat,Monkey
Conjugate: unconjugated